REPLICATION
The proposal by James Watson and Francis Crick in 1953, which included the concept of DNA replication, was a pivotal moment in the understanding of genetics and molecular biology. Their statement, “It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material,” was a key insight that laid the foundation for our understanding of DNA replication.
The scheme they proposed, known as semiconservative DNA replication, can be summarized as follows:
Separation of Strands: The two strands of the DNA double helix would separate, effectively “unzipping” the double helix. This separation would be facilitated by breaking the hydrogen bonds between the complementary base pairs (A with T, and G with C).
Template for Synthesis: Each separated strand would then serve as a template for the synthesis of a new complementary strand. This means that for each original DNA strand, a new DNA strand would be synthesized according to the base-pairing rules: A with T and G with C.
Complementary Base Pairing: During the synthesis of the new strand, free nucleotides in the cell’s environment would be brought in and added to the growing strand. Adenine (A) would pair with thymine (T), and guanine (G) would pair with cytosine (C).
Formation of New DNA Molecule: After replication is completed, each DNA molecule would consist of one original parental strand and one newly synthesized strand. These two strands would be complementary to each other.
The concept of semiconservative DNA replication was later confirmed through experiments, most notably by Matthew Meselson and Franklin Stahl in 1958, using a technique called density gradient centrifugation. Their experiment provided direct evidence that DNA replication follows the semiconservative model.
Matthew Meselson and Franklin Stahl’s experiment, conducted in 1958, provided concrete evidence for the semiconservative replication of DNA. This landmark experiment used the bacterium Escherichia coli (E. coli) and the heavy isotope of nitrogen (15N) to track the replication of DNA. Here’s an overview of the experiment:
Objective:
- The primary objective of the experiment was to determine the mode of DNA replication (conservative, semiconservative, or dispersive) in E. coli.
Experimental Procedure:
Isotopic Labeling: E. coli bacteria were grown in a medium containing 15NH4Cl, where 15N (a heavy isotope of nitrogen) was the sole source of nitrogen for many generations. This heavy nitrogen isotope was incorporated into nitrogen-containing compounds in the cells, including DNA.
Transfer to Normal Medium: After multiple generations in the 15N medium, the E. coli cells were transferred to a medium containing normal 14NH4Cl, which contained the lighter isotope of nitrogen (14N).
Sampling and DNA Extraction: At various time intervals following the transfer to the normal medium, samples of E. coli cells were taken as they multiplied. DNA was extracted from these samples.
Centrifugation in CsCl Gradient: The extracted DNA from each sample was subjected to centrifugation in a cesium chloride (CsCl) density gradient. The CsCl gradient allowed the separation of DNA molecules based on their density. Heavier DNA molecules would settle lower in the gradient, while lighter DNA molecules would stay higher.
Key Observations:
- The DNA extracted from the E. coli culture one generation (20 minutes) after the transfer from 15N to 14N medium had an intermediate density. This intermediate density indicated that it was a hybrid molecule consisting of one heavy strand (15N-labeled) and one light strand (14N-labeled).
- The DNA extracted from the E. coli culture after an additional generation (40 minutes, 2nd generation) was composed of equal amounts of the hybrid DNA and “light” DNA, which had the same density as DNA from cells grown entirely in 14N medium.
Conclusions:
- The experiment’s results supported the semiconservative replication model proposed by Watson and Crick. The presence of DNA molecules with intermediate density indicated that one strand of the DNA helix was conserved and used as a template for the synthesis of a complementary, light DNA strand.
- After one round of replication (one generation), the DNA in the E. coli population became a mix of hybrid DNA (one heavy strand and one light strand) and light DNA (two light strands).
- Subsequent generations would result in a higher proportion of light DNA, consistent with semiconservative replication.
Significance:
- The Meselson-Stahl experiment provided strong empirical evidence for the semiconservative replication of DNA, confirming Watson and Crick’s proposal.
- It is considered one of the most iconic experiments in molecular biology and has had a profound impact on our understanding of DNA replication and inheritance.
The experiments conducted by Taylor and colleagues on Vicia faba (faba beans) in 1958, involving the use of radioactive thymidine to detect the distribution of newly synthesized DNA in the chromosomes, provided further evidence supporting the semiconservative replication of DNA. These experiments were consistent with the earlier findings of Matthew Meselson and Franklin Stahl, which were conducted using Escherichia coli.
In the experiments on Vicia faba, radioactive thymidine (a precursor to DNA) was incorporated into newly synthesized DNA during replication. This radioactive label allowed researchers to track the distribution of newly synthesized DNA in the chromosomes.
The key observations and conclusions from these experiments were likely similar to those of the Meselson-Stahl experiment:
Distribution of Radioactive DNA: When radioactive thymidine was incorporated into the newly synthesized DNA, the radiolabeled DNA was distributed within the chromosomes.
Evidence of Semiconservative Replication: The distribution of radiolabeled DNA within the chromosomes supported the semiconservative replication model. If DNA replication were conservative or dispersive, the distribution of radiolabeled DNA in the chromosomes would have been different.
These experiments on Vicia faba provided independent confirmation of the semiconservative replication of DNA, reinforcing the understanding that DNA replication occurs in a semiconservative manner, where each newly synthesized DNA molecule contains one parental (old) strand and one newly synthesized strand.
The Machinery and the Enzymes
DNA replication is a highly complex and tightly regulated process in living cells, involving a set of enzymes that work together to ensure the accurate duplication of genetic material. Here’s an overview of the key enzymes and processes involved in DNA replication:
DNA-Dependent DNA Polymerase: The main enzyme responsible for DNA replication is DNA-dependent DNA polymerase. This enzyme catalyzes the polymerization of deoxynucleotides to form a new DNA strand based on a DNA template. It works in the 5′ to 3′ direction, meaning it adds nucleotides to the growing strand in the 3′ to 5′ direction of the template strand. DNA polymerases are highly efficient and have to work rapidly to replicate the entire genome. In E. coli, for example, replication is completed in just 18 minutes.
Accuracy of DNA Polymerase: DNA polymerases must work with high fidelity to minimize errors during replication. Any mistake could lead to mutations, which may have detrimental effects. These enzymes have proofreading mechanisms to correct errors, and they have error rates as low as one error in every billion base pairs replicated.
Energy Source: The energy required for polymerization comes from the deoxyribonucleoside triphosphates (dNTPs) that serve as both substrates and energy sources for the reaction. The hydrolysis of the high-energy phosphate bonds in dNTPs provides the energy needed for the polymerization process.
Additional Enzymes: DNA replication involves several other enzymes to support the process. These include DNA helicase, which unwinds the DNA double helix to create a replication fork, and DNA primase, which synthesizes short RNA primers on the DNA template to initiate DNA synthesis.
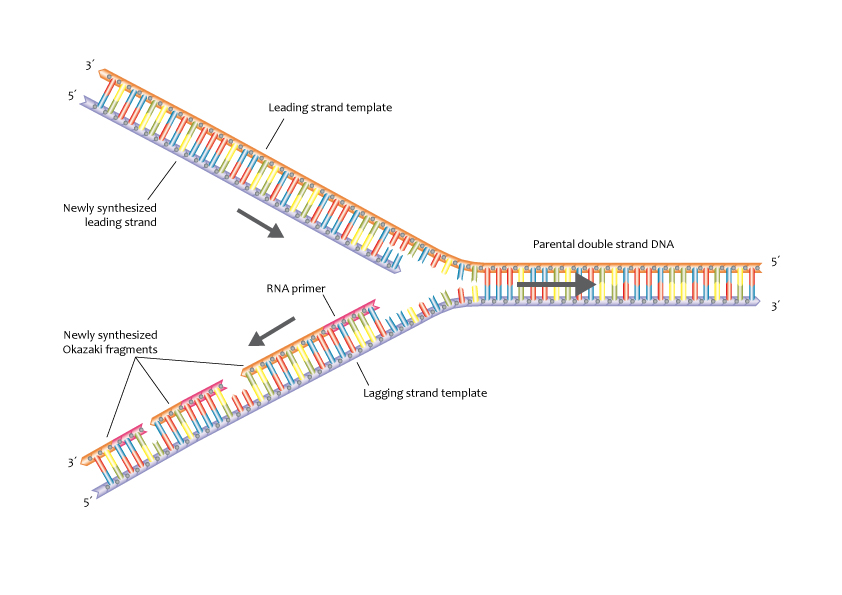
Replication Fork: DNA replication occurs at a replication fork, which is a small opening in the DNA helix. One strand, called the leading strand, is replicated continuously, while the other strand, the lagging strand, is replicated discontinuously in the form of short fragments. These fragments are known as Okazaki fragments.
DNA Ligase: DNA ligase is responsible for joining the Okazaki fragments on the lagging strand, sealing any nicks or gaps in the newly synthesized DNA.
Origin of Replication: DNA replication begins at specific regions in the DNA known as the origin of replication. In E. coli, for instance, there is a well-defined origin of replication that serves as the starting point for replication.
Replication Direction: DNA polymerases work in the 5′ to 3′ direction, creating some complexities at the replication fork. The leading and lagging strands are synthesized in opposite directions due to this directional constraint.
Vectors in DNA Propagation: Vectors, which are often used in recombinant DNA procedures, contain their own origin of replication. This is necessary for propagating the DNA of interest and ensuring its replication within host cells.